KNN vs PNN Classification: Breast Cancer Image Dataset¶
In addition to powerful manifold learning and network graphing algorithms, the SliceMatrix-IO platform contains serveral classification algorithms. Classification is one of the foundational tasks of machine learning: given an input data vector, a classifier attempts to guess the correct class label. Today we will look at two supervised classifiers: the K-Nearest Neighbor Classifier and Probabilistic Neural Network Classifier.
Features are computed from a digitized image of a fine needle aspirate (FNA) of a breast mass. They describe characteristics of the cell nuclei present in the image. For example:
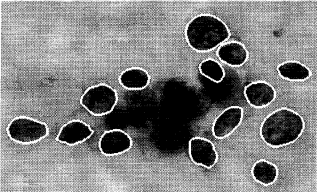
Attributes:
- Sample code number: id number
- Clump Thickness: 1 - 10
- Uniformity of Cell Size: 1 - 10
- Uniformity of Cell Shape: 1 - 10
- Marginal Adhesion: 1 - 10
- Single Epithelial Cell Size: 1 - 10
- Bare Nuclei: 1 - 10
- Bland Chromatin: 1 - 10
- Normal Nucleoli: 1 - 10
- Mitoses: 1 - 10
- Class: (2 for benign, 4 for malignant)
The goal in this example is to use the input features to train a machine learning model to predict whether each image presented is benign or malignant. We will then validate the predictive power of the model using out of sample data.
To do this, we begin by importing the SliceMatrix-IO Python client.
If you haven't installed the client yet, the easiest way is with pip:
pip install slicematrixIO
Next, lets import slicematrixIO and create our client which will do the heavy lifting. Make sure to replace the api key below with your own key.
Don't have a key yet? Get your api key here
from slicematrixIO import SliceMatrix
api_key = "insert your api key here"
sm = SliceMatrix(api_key)
To begin, let's import some useful libraries
import pandas as pd
import numpy as np
np.random.seed(98765) #reproducability
Let's load the full dataset...
training_data = pd.read_csv("notebook_files/breast-cancer-wisconsin.data",index_col = 0, header = None)
training_data = training_data[training_data.ix[:,6] != '?']
training_data.index = np.arange(0,training_data.shape[0], 1)
...then shuffle and split the data into training and testing sets...
shuffled = training_data.reindex(np.random.permutation(training_data.index))
shuffled.index = np.arange(0, training_data.shape[0], 1)
cols = shuffled.columns.values.tolist()
cols[-1] = "class"
shuffled.columns = cols
data = shuffled.ix[0:training_data.shape[0]/2,:]
out = shuffled.ix[training_data.shape[0]/2:,:]
data.head()
knn = sm.KNNClassifier(dataset = data, class_column = "class")
Now we can make predictions using the out of sample (testing / validation) data
validation_preds = knn.predict(out.drop("class",axis =1).values.tolist())
Finally we can calculate the percentage of out of sample predictions which were correct...
pct_correct = 1. * np.sum(np.equal(validation_preds, out['class'])) / len(validation_preds)
print pct_correct
In addition, we could compare performance against another classifier: the Probabilistic Neural Network
pnn = sm.PNNClassifier(dataset = data, class_column = "class")
validation_preds2 = pnn.predict(out.drop("class",axis =1).values.tolist())
pct_correct2 = 1. * np.sum(np.equal(validation_preds2, out['class'])) / len(validation_preds)
print pct_correct2
Don't have a SliceMatrix-IO api key yet? Get your api key here
